Vitamin D - A master example of nutrigenomics
Vitamin D - A master example of nutrigenomics
Redox Biology https://doi.org/10.1016/j.redox.2023.102695
Carsten Carlberg a b, Marianna Raczyk a, Natalia Zawrotna a
Nutrigenomics attempts to characterize and integrate the relation between dietary molecules and gene expression on a genome-wide level. One of the biologically active nutritional compounds is vitamin D3, which activates via its metabolite 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3) the nuclear receptor VDR (vitamin D receptor). Vitamin D3 can be synthesized endogenously in our skin, but since we spend long times indoors and often live at higher latitudes where for many winter months UV-B radiation is too low, it became a true vitamin.
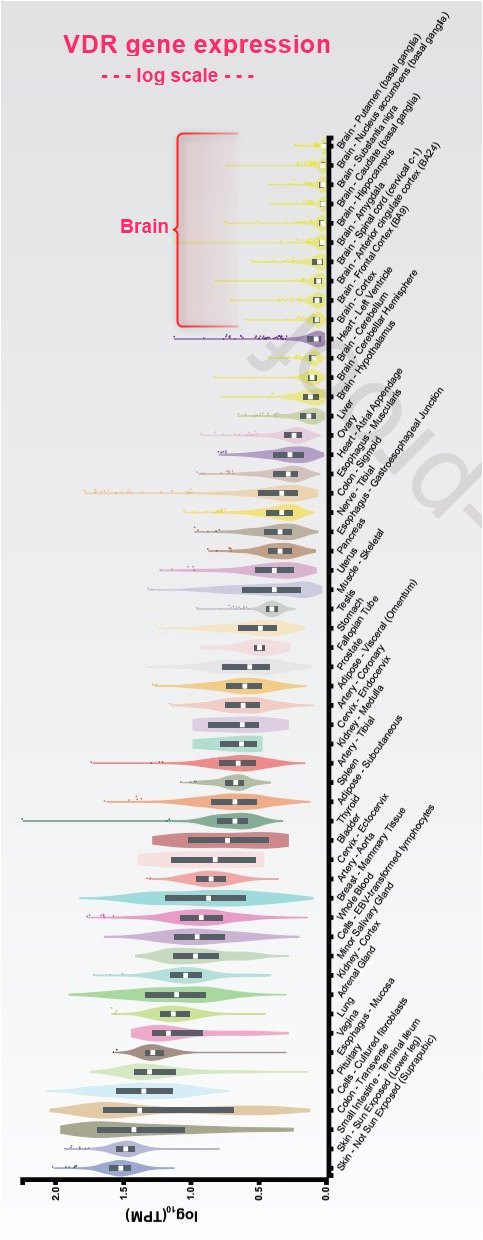
The ligand-inducible transcription factor VDR is expressed in the majority of human tissues and cell types, where it modulates the epigenome at thousands of genomic sites. In a tissue-specific fashion this results in the up- and downregulation of primary vitamin D target genes, some of which are involved in attenuating oxidative stress.
Vitamin D affects a wide range of physiological functions including the control of metabolism, bone formation and immunity. In this review, we will discuss how the epigenome- and transcriptome-wide effects of 1,25(OH)2D3 and its receptor VDR serve as a master example in nutrigenomics. In this context, we will outline the basis of a mechanistic understanding for personalized nutrition with vitamin D3.
📄 Download the PDF from Vitamin D Life
References (34 = Carlberg)
Carlberg , C., Ulven, S. M. & Molnar, F. Nutrigenomics: how science works. Springer textbook, doi:10.1007/978-3-030-36948-4 (2020).
Meyer, C. A. & Liu, X. S. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet 15, 709-721, doi:10.1038/nrg3788 (2014).
Ferguson, J. F. et al. Nutrigenomics, the microbiome, and gene-environment interactions: new directions in cardiovascular disease research, prevention, and treatment: a scientific statement from the American Heart Association. Circ Cardiovasc Genet 9, 291-313, doi:10.1161/HCG.0000000000000030 (2016).
Fenech, M. et al. Nutrigenetics and Nutrigenomics: viewpoints on the current status and applications in nutrition research and practice. Journal of nutrigenetics and nutrigenomics 4, 69-89, doi:10.1159/000327772 (2011).
- Journal has 347 papers as of April 2023
Müller, M. & Kersten, S. Nutrigenomics: goals and strategies. Nat Rev Genet 4, 315322, doi:10.1038/nrg1047 (2003).
Vanden Berghe, W. Epigenetic impact of dietary polyphenols in cancer chemoprevention: lifelong remodeling of our epigenomes. Pharmacological research : the official journal of the Italian Pharmacological Society 65, 565-576, doi:10.1016/j.phrs.2012.03.007 (2012).
Bendik, I., Friedel, A., Roos, F. F., Weber, P. & Eggersdorfer, M. Vitamin D: a critical and essential micronutrient for human health. Front Physiol 5, 248, doi:10.3389/fphys.2014.00248 (2014).
Carlberg , C. Molecular endocrinology of vitamin D on the epigenome level. Mol Cell Endocrinol 453, 14-21, doi:10.1016/j.mce.2017.03.016 (2017).
Hanel, A. & Carlberg , C. Time-resolved gene expression analysis monitors the regulation of inflammatory mediators and attenuation of adaptive immune response by vitamin D. Int J Mol Sci 23, doi:10.3390/ijms23020911 (2022).
Malmberg, H. R., Hanel, A., Taipale, M., Heikkinen, S. & Carlberg , C. Vitamin D treatment sequence is critical for transcriptome modulation of immune challenged primary human cells. Frontiers in immunology 12, 754056, doi:10.3389/fimmu.2021.754056 (2021).
Sosa-Diaz, E., Hernandez-Cruz, E. Y. & Pedraza-Chaverri, J. The role of vitamin D on redox regulation and cellular senescence. Free Radic Biol Med 193, 253-273, doi:10.1016/j.freeradbiomed.2022.10.003 (2022).
Tremezaygues, L. et al. Cutaneous photosynthesis of vitamin D: an evolutionary highly-conserved endocrine system that protects against environmental hazards including UV-radiation and microbial infections. Anticancer Res 26, 2743-2748 (2006).
Holick, M. F. Photobiology of vitamin D. Vitamin D 3rd edn, 13-22, doi:10.1016/b978-0-12-381978-9.10002-2 (2011).
Capell-Hattam, I. M. & Brown, A. J. Sterol evolution: cholesterol synthesis in animals is less a required trait than an acquired taste. Curr Biol 30, R886-R888, doi:10.1016/j.cub.2020.06.007 (2020).
Jasinghe, V. J., Perera, C. O. & Barlow, P. J. Bioavailability of vitamin D2 from irradiated mushrooms: an in vivo study. Br JNutr 93, 951-955 (2005).
Holick, M. F. Sunlight and vitamin D for bone health and prevention of autoimmune diseases, cancers, and cardiovascular disease. Am J Clin Nutr 80, 1678S-1688S (2004).
Bouillon, R. & Suda, T. Vitamin D: calcium and bone homeostasis during evolution. BoneKEy Reports 3, 480, doi:10.1038/bonekey.2013.214 (2014).
Tripkovic, L. et al. Comparison of vitamin D2 and vitamin D3 supplementation in raising serum 25-hydroxyvitamin D status: a systematic review and meta-analysis. Am JClin Nutr 95, 1357-1364, doi:10.3945/ajcn.111.031070 (2012).
Lamberg-Allardt, C. Vitamin D in foods and as supplements. Progress in biophysics and molecular biology 92, 33-38, doi:10.1016/j.pbiomolbio.2006.02.017 (2006).
Bikle, D. D. & Schwartz, J. Vitamin D binding protein, total and free vitamin D levels in different physiological and pathophysiological conditions. Front Endocrinol 10, 317, doi:10.3389/fendo.2019.00317 (2019).
Bikle, D. D. Vitamin D metabolism, mechanism of action, and clinical applications. Chem Biol 21, 319-329, doi:10.1016/j.chembiol.2013.12.016 (2014).
Norman, A. W. From vitamin D to hormone D: fundamentals of the vitamin D endocrine system essential for good health. Am J Clin Nutr 88, 491S-499S (2008).
Bikle, D. & Christakos, S. New aspects of vitamin D metabolism and action - addressing the skin as source and target. Nat Rev Endocrinol, doi:10.1038/s41574-019- 0312-5 (2020).
Haussler, M. R., Jurutka, P. W., Mizwicki, M. & Norman, A. W. Vitamin D receptor (VDR)-mediated actions of 1a,25(OH)2vitamin D3: genomic and non-genomic mechanisms. Best practice & research. Clinical endocrinology & metabolism 25, 543559, doi:10.1016/j.beem.2011.05.010 (2011).
Hewison, M. et al. Extra-renal 25-hydroxyvitamin D3-1a-hydroxylase in human health and disease. J Steroid Biochem Mol Biol 103, 316-321, doi:10.1016/j.jsbmb.2006.12.078 (2007).
Zerwekh, J. E. Blood biomarkers of vitamin D status. Am J Clin Nutr 87, 1087S-1091S, doi:10.1093/ajcn/87.4.1087S (2008).
Hollis, B. W. Circulating 25-hydroxyvitamin D levels indicative of vitamin D sufficiency: implications for establishing a new effective dietary intake recommendation for vitamin D. J Nutr 135, 317-322 (2005).
Institute-of-Medicine. Dietary reference intakes for calcium and vitamin D. Washington, DC: National Academies Press (2011).
Bouillon, R. et al. Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr Rev 29, 726-776 (2008).
Sintzel, M. B., Rametta, M. & Reder, A. T. Vitamin D and multiple sclerosis: a comprehensive review. Neurol Ther 7, 59-85, doi:10.1007/s40120-017-0086-4 (2018).
Ramagopalan, S. V. et al. Expression of the multiple sclerosis-associated MHC class II Allele HLA-DRB1*1501 is regulated by vitamin D. PLoS genetics 5, e1000369, doi:10.1371/journal.pgen.1000369 (2009).
Jeffery, L. E., Raza, K. & Hewison, M. Vitamin D in rheumatoid arthritis-towards clinical application. Nat Rev Rheumatol 12, 201-210, doi:10.1038/nrrheum.2015.140 (2016).
Fletcher, J., Cooper, S. C., Ghosh, S. & Hewison, M. The role of vitamin D in Inflammatory bowel disease: mechanism to management. Nutrients 11, doi:10.3390/nu11051019 (2019).
Infante, M. et al. Influence of vitamin D on islet autoimmunity and beta-cell function in type 1 diabetes. Nutrients 11, doi:10.3390/nu11092185 (2019).
Huang, S. J. et al. Vitamin D deficiency and the risk of tuberculosis: a meta-analysis. DrugDesDevel Ther 11, 91-102, doi:10.2147/DDDT.S79870 (2017).
Rook, G. A. The role of vitamin D in tuberculosis. Am Rev Respir Dis 138, 768-770, doi:10.1164/ajrccm/138.4.768 (1988).
Charoenngam, N., Shirvani, A. & Holick, M. F. Vitamin D and Its potential benefit for the COVID-19 pandemic. Endocrine practice : officialjournal of the American College of Endocrinology and the American Association of Clinical Endocrinologists 27, 484493, doi:10.1016/j.eprac.2021.03.006 (2021).
Maghbooli, Z. et al. Vitamin D sufficiency, a serum 25-hydroxyvitamin D at least 30 ng/mL reduced risk for adverse clinical outcomes in patients with COVID-19 infection. PLoS ONE 15, e0239799, doi:10.1371/journal.pone.0239799 (2020).
Pludowski, P. et al. Vitamin D effects on musculoskeletal health, immunity, autoimmunity, cardiovascular disease, cancer, fertility, pregnancy, dementia and mortality-a review of recent evidence. Autoimmun Rev 12, 976-989, doi:10.1016/j.autrev.2013.02.004 (2013).
Carlberg , C. Molecular approaches for optimizing vitamin D supplementation. Vitamins and hormones 100, 255-271, doi:10.1016/bs.vh.2015.10.001 (2016).
Lu, Z. et al. An evaluation of the vitamin D3 content in fish: Is the vitamin D content adequate to satisfy the dietary requirement for vitamin D? J Steroid Biochem Mol Biol 103, 642-644, doi:10.1016/j.jsbmb.2006.12.010 (2007).
Urbain, P., Singler, F., Ihorst, G., Biesalski, H. K. & Bertz, H. Bioavailability of vitamin D2 from UV-B-irradiated button mushrooms in healthy adults deficient in serum 25- hydroxyvitamin D: a randomized controlled trial. European journal of clinical nutrition 65, 965-971, doi:10.1038/ejcn.2011.53 (2011).
Holick, M. F. et al. Evaluation, treatment, and prevention of vitamin D deficiency: an Endocrine Society clinical practice guideline. J Clin EndocrinolMetab 96, 1911-1930, doi:10.1210/jc.2011-0385 (2011).
Cheskis, B. J., Freedman, L. P. & Nagpal, S. Vitamin D receptor ligands for osteoporosis. Curr Opin InvestigDrugs 7, 906-911 (2006).
Krasowski, M. D., Ni, A., Hagey, L. R. & Ekins, S. Evolution of promiscuous nuclear hormone receptors: LXR, FXR, VDR, PXR, and CAR. Mol Cell Endocrinol 334, 3948, doi:10.1016/j.mce.2010.06.016 (2011).
Makishima, M. et al. Vitamin D receptor as an intestinal bile acid sensor. Science 296, 1313-1316, doi:10.1126/science.1070477 (2002).
Makishima, M. et al. Identification of a nuclear receptor for bile acids. Science 284, 1362-1365, doi:10.1126/science.284.5418.1362 (1999).
Staudinger, J. L. et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 98, 3369-3374., doi:10.1073/pnas.051551698 (2001).
Guo, G. L. et al. Complementary roles of farnesoid X receptor, pregnane X receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem 278, 45062-45071, doi:10.1074/jbc.M307145200 (2003).
Whitfield, G. K. et al. Cloning of a functional vitamin D receptor from the lamprey (Petromyzon marinus), an ancient vertebrate lacking a calcified skeleton and teeth. Endocrinology 144, 2704-2716, doi:10.1210/en.2002-221101 (2003).
Hanel, A. & Carlberg , C. Vitamin D and evolution: pharmacologic implications. Biochem Pharmacol 173, 113595, doi:10.1016/j.bcp.2019.07.024 (2020).
Meyer, M. B. & Pike, J. W. Genomic mechanisms controlling renal vitamin D metabolism. J Steroid Biochem Mol Biol 228, 106252, doi:10.1016/j.jsbmb.2023.106252 (2023).
Heikkinen, S. et al. Nuclear hormone 1a,25-dihydroxyvitamin D3 elicits a genomewide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res 39, 91819193, doi:10.1093/nar/gkr654 (2011).
Vanherwegen, A. S. et al. Vitamin D controls the capacity of human dendritic cells to induce functional regulatory T cells by regulation of glucose metabolism. J Steroid Biochem Mol Biol 187, 134-145, doi:10.1016/j.jsbmb.2018.11.011 (2019).
Vanherwegen, A. S., Gysemans, C. & Mathieu, C. Vitamin D endocrinology on the cross-road between immunity and metabolism. Mol Cell Endocrinol 453, 52-67, doi:10.1016/j.mce.2017.04.018 (2017).
Chun, R. F., Liu, P. T., Modlin, R. L., Adams, J. S. & Hewison, M. Impact of vitamin D on immune function: lessons learned from genome-wide analysis. Front Physiol 5, 151, doi:10.3389/fphys.2014.00151 (2014).
Gombart, A. F. The vitamin D-antimicrobial peptide pathway and its role in protection against infection. Future Microbiol 4, 1151-1165, doi:10.2217/fmb.09.87 (2009).
Zanoni, I. & Granucci, F. Role of CD14 in host protection against infections and in metabolism regulation. Front Cell Infect Microbiol 3, 32, doi:10.3389/fcimb.2013.00032 (2013).
Lu, M., McComish, B. J., Burdon, K. P., Taylor, B. V. & Korner, H. The association between vitamin D and multiple sclerosis risk: 1,25(OH)2D3 induces super-enhancers bound by VDR. Frontiers in immunology 10, 488, doi:10.3389/fimmu.2019.00488 (2019).
Dankers, W., Colin, E. M., van Hamburg, J. P. & Lubberts, E. Vitamin D in autoimmunity: molecular mechanisms and therapeutic potential. Frontiers in immunology 7, 697, doi:10.3389/fimmu.2016.00697 (2016).
Carlberg , C. & Velleuer, E. Molecular immunology: how science works. Springer Textbook (2022).
Zanoni, I., Tan, Y., Di Gioia, M., Springstead, J. R. & Kagan, J. C. By capturing inflammatory lipids released from dying cells, the receptor CD14 induces inflammasome-dependent phagocyte hyperactivation. Immunity 47, 697-709 e693, doi:10.1016/j.immuni.2017.09.010 (2017).
Hoeksema, M. A. & de Winther, M. P. Epigenetic regulation of monocyte and macrophage function. AntioxidRedox Signal 25, 758-774, doi:10.1089/ars.2016.6695 (2016).
Liang, S., Cai, J., Li, Y. & Yang, R. 1,25Dihydroxyvitamin D3 induces macrophage polarization to M2 by upregulating T cell Igmucin 3 expression. Mol Med Rep 19, 37073713, doi:10.3892/mmr.2019.10047 (2019).
Novershtern, N. et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296-309, doi:10.1016/j.cell.2011.01.004 (2011).
Cortes, M. et al. Developmental vitamin D availability impacts hematopoietic stem cell production. Cell reports 17, 458-468, doi:10.1016/j.celrep.2016.09.012 (2016).
Koivisto, O., Hanel, A. & Carlberg , C. Key vitamin D target genes with functions in the immune system. Nutrients 12, doi:10.3390/nu12041140 (2020).
Zeitelhofer, M. et al. Functional genomics analysis of vitamin D effects on CD4+ T cells in vivo in experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 114, E1678-E1687, doi:10.1073/pnas.1615783114 (2017).
Barragan, M., Good, M. & Kolls, J. K. Regulation of dendritic cell function by vitamin D. Nutrients 7, 8127-8151, doi:10.3390/nu7095383 (2015).
Sever, R. & Brugge, J. S. Signal transduction in cancer. Cold Spring Harb Perspect Med 5, doi:10.1101/cshperspect.a006098 (2015).
Meyer, M. B., Goetsch, P. D. & Pike, J. W. VDR/RXR and TCF4/beta-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol Endocrinol 26, 37-51, doi:10.1210/me.2011-1109 (2012).
Palmer, H. G. et al. Genetic signatures of differentiation induced by 1a,25- dihydroxyvitamin D3 in human colon cancer cells. Cancer Res 63, 7799-7806 (2003).
Wood, R. J., Tchack, L., Angelo, G., Pratt, R. E. & Sonna, L. A. DNA microarray analysis of vitamin D-induced gene expression in a human colon carcinoma cell line. Physiol Genomics 17, 122-129, doi:10.1152/physiolgenomics.00002.2003 (2004).
Salehi-Tabar, R. et al. Vitamin D receptor as a master regulator of the c-MYC/MXD1 network. Proc Natl Acad Sci U S A 109, 18827-18832, doi:10.1073/pnas.1210037109 (2012).
Sinkkonen, L., Malinen, M., Saavalainen, K., Vaisanen, S. & Carlberg , C. Regulation of the human cyclin C gene via multiple vitamin D3-responsive regions in its promoter. Nucleic Acids Res 33, 2440-2451, doi:10.1093/nar/gki502 (2005).
Saramaki, A., Banwell, C. M., Campbell, M. J. & Carlberg , C. Regulation of the human p21waf1/cip1 gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res 34, 543-554, doi:10.1093/nar/gkj460 (2006).
Saramaki, A. et al. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1a,25- dihydroxyvitamin D3. J Biol Chem 284, 8073-8082, doi:10.1074/jbc.M808090200 (2009).
Toropainen, S., Vaisanen, S., Heikkinen, S. & Carlberg , C. The down-regulation of the human MYC gene by the nuclear hormone 1a,25-dihydroxyvitamin D3 is associated with cycling of corepressors and histone deacetylases. J Mol Biol 400, 284-294, doi:10.1016/j.jmb.2010.05.031 (2010).
Abe, E. et al. Differentiation of mouse myeloid leukemia cells induced by 1a,25- dihydroxyvitamin D3. Proc Natl Acad Sci USA 78, 4990-4994 (1981).
Colston, K., Colston, M. J., Fieldsteel, A. H. & Feldman, D. 1,25-dihydroxyvitamin D3 receptors in human epithelial cancer cell lines. Cancer Research 42, 856-859 (1982).
Balomenos, D. et al. The cell cycle inhibitor p21 controls T-cell proliferation and sex- linked lupus development. Nat Med 6, 171-176, doi:10.1038/72272 (2000).
Martinez-Lostao, L., Anel, A. & Pardo, J. How do cytotoxic lymphocytes kill cancer cells? Clin Cancer Res 21, 5047-5056, doi:10.1158/1078-0432.CCR-15-0685 (2015).
Doherty, A. H., Ghalambor, C. K. & Donahue, S. W. Evolutionary physiology of bone: bone metabolism in changing environments. Physiology (Bethesda) 30, 17-29, doi:10.1152/physiol.00022.2014 (2015).
van de Peppel, J. & van Leeuwen, J. P. Vitamin D and gene networks in human osteoblasts. Front Physiol 5, 137, doi:10.3389/fphys.2014.00137 (2014).
van Driel, M. & van Leeuwen, J. Vitamin D and Bone: A Story of Endocrine and Auto/Paracrine Action in Osteoblasts. Nutrients 15, doi:10.3390/nu15030480 (2023).
Latic, N. & Erben, R. G. Interaction of vitamin D with peptide hormones with emphasis on parathyroid hormone, FGF23, and the renin-angiotensin-aldosterone system. Nutrients 14, doi:10.3390/nu14235186 (2022).
Veldurthy, V. et al. Vitamin D, calcium homeostasis and aging. Bone Res 4, 16041, doi:10.1038/boneres.2016.41 (2016).
Bar, L. et al. Insulin suppresses the production of fibroblast growth factor 23 (FGF23). Proc Natl Acad Sci U S A 115, 5804-5809, doi:10.1073/pnas.1800160115 (2018).
Hanel, A. et al. Common and personal target genes of the micronutrient vitamin D in primary immune cells from human peripheral blood. Scientific reports 10, 21051, doi:10.1038/s41598-020-78288-0 (2020).
McCarthy, K. et al. Association between vitamin D deficiency and the risk of prevalent type 2 diabetes and incident prediabetes: A prospective cohort study using data from The Irish Longitudinal Study on Ageing (TILDA). EClinicalMedicine 53, 101654, doi:10.1016/j.eclinm.2022.101654 (2022).
Melguizo-Rodriguez, L. et al. Role of Vitamin D in the Metabolic Syndrome. Nutrients 13, doi:10.3390/nu13030830 (2021).
Kim-Hellmuth, S. et al. Cell type-specific genetic regulation of gene expression across human tissues. Science 369, doi:10.1126/science.aaz8528 (2020).
Carlberg , C. & Molnar, F. The impact of chromatin. Mechanisms of Gene Regulation, 2nded., Springer, 17-34, doi:DOI: 10.1007/978-94-017-7741-4_2 (2016).
Michael, A. K. & Thoma, N. H. Reading the chromatinized genome. Cell 184, 35993611, doi:10.1016/j.cell.2021.05.029 (2021).
Beisel, C. & Paro, R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet 12, 123-135, doi:10.1038/nrg2932 (2011).
Rivera, Chloe M. & Ren, B. Mapping human epigenomes. Cell 155, 39-55, doi:10.1016/j.cell.2013.09.011 (2013).
Hathaway, N. A. et al. Dynamics and memory of heterochromatin in living cells. Cell 149, 1447-1460, doi:10.1016/j.cell.2012.03.052 (2012).
Perino, M. & Veenstra, G. J. Chromatin control of developmental dynamics and plasticity. Dev Cell 38, 610-620, doi:10.1016/j.devcel.2016.08.004 (2016).
Carlberg , C. & Molnar, F. The epigenome. Mechanisms of Gene Regulation, 2nd ed., Springer, 159-172, doi:DOI: 10.1007/978-94-017-7741-4_8 (2016).
Li, J. et al. Metabolic control of histone acetylation for precise and timely regulation of minor ZGA in early mammalian embryos. CellDiscov 8, 96, doi:10.1038/s41421-022- 00440-z (2022).
Su, C. et al. 3D chromatin maps of the human pancreas reveal lineage-specific regulatory architecture of T2D risk. Cell Metab 34, 1394-1409 e1394, doi:10.1016/j.cmet.2022.08.014 (2022).
Bruno, S., Williams, R. J. & Del Vecchio, D. Epigenetic cell memory: the gene's inner chromatin modification circuit. PLoS computational biology 18, e1009961, doi:10.1371/journal.pcbi.1009961 (2022).
Fitz-James, M. H. & Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat Rev Genet 23, 325-341, doi:10.1038/s41576-021-00438-5 (2022).
Carlberg , C. & Molnar, F. Chromatin modifiers. Mechanisms of Gene Regulation, 2nd ed., Springer, 129-145, doi:DOI: 10.1007/978-94-017-7741-4_8 (2016).
Gut, P. & Verdin, E. The nexus of chromatin regulation and intermediary metabolism. Nature 502, 489-498, doi:10.1038/nature12752 (2013).
Chen, C., Wang, Z. & Qin, Y. Connections between metabolism and epigenetics: mechanisms and novel anti-cancer strategy. Frontiers in pharmacology 13, 935536, doi:10.3389/fphar.2022.935536 (2022).
Dai, Z., Ramesh, V. & Locasale, J. W. The evolving metabolic landscape of chromatin biology and epigenetics. Nat Rev Genet 21, 737-753, doi:10.1038/s41576-020-0270-8 (2020).
Carlberg , C., Ulven, S. M. & Molnar, F. Nutrigenomics. Springer Textbook (2016).
Carlberg , C. & Molnar, F. Human epigenomics, Springer (2018).
Pike, J. W. et al. Perspectives on mechanisms of gene regulation by 1,25- dihydroxyvitamin D3 and its receptor. J Steroid Biochem Mol Biol 103, 389-395 (2007).
Mangelsdorf, D. J. & Evans, R. M. The RXR heterodimers and orphan receptors. Cell 83, 841-850 (1995).
Carlberg , C. Genome-wide (over)view on the actions of vitamin D. Front Physiol 5, 167, doi:10.3389/fphys.2014.00167 (2014).
Ramagopalan, S. V. et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome research 20, 1352-1360 (2010).
Neme, A., Seuter, S. & Carlberg , C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim Biophys Acta 1860, 952-961, doi:10.1016/j.bbagrm.2017.07.002 (2017).
Ding, N. et al. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell 153, 601-613, doi:10.1016/j.cell.2013.03.028 (2013).
Tuoresmaki, P., Vaisanen, S., Neme, A., Heikkinen, S. & Carlberg , C. Patterns of
genome-wide VDR locations. PLoS ONE 9, e96105, doi:10.1371/journal.pone.0096105 (2014).
Molnár, F., Perakyla, M. & Carlberg , C. Vitamin D receptor agonists specifically modulate the volume of the ligand-binding pocket. J Biol Chem 281, 10516-10526, doi:10.1074/jbc.M513609200 (2006).
Polly, P. et al. VDR-Alien: a novel, DNA-selective vitamin D3 receptor-corepressor partnership. Faseb J 14, 1455-1463 (2000).
Herdick, M. & Carlberg , C. Agonist-triggered modulation of the activated and silent state of the vitamin D3 receptor by interaction with co-repressors and co-activators. J. Mol. Biol. 304, 793-801 (2000).
Nurminen, V., Neme, A., Seuter, S. & Carlberg , C. The impact of the vitamin D- modulated epigenome on VDR target gene regulation. Biochim Biophys Acta 1861, 697-705, doi:10.1016/j.bbagrm.2018.05.006. (2018).
Nurminen, V., Neme, A., Seuter, S. & Carlberg , C. Modulation of vitamin D signaling by the pioneer factor CEBPA. Biochim Biophys Acta 1862, 96-106, doi:10.1016/j.bbagrm.2018.12.004 (2019).
Seuter, S., Neme, A. & Carlberg , C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res 44, 4090-4104, doi:10.1093/nar/gkv1519 (2016).
Zaret, K. S. & Carroll, J. S. Pioneer transcription factors: establishing competence for gene expression. Gen Dev 25, 2227-2241 (2011).
Seuter, S., Neme, A. & Carlberg , C. Epigenomic PU.1-VDR crosstalk modulates vitamin D signaling. Biochim Biophys Acta 1860, 405-415, doi:10.1016/j.bbagrm.2017.02.005 (2017).
Seuter, S., Neme, A. & Carlberg , C. ETS transcription factor family member GABPA contributes to vitamin D receptor target gene regulation. J Steroid Biochem Mol Biol 177, 46-52, doi:10.1016/j.jsbmb.2017.08.006 (2018).
Carlberg , C. Vitamin D and is target genes. Nutrients 14, doi:10.3390/nu14071354 (2022).
Saccone, D., Asani, F. & Bornman, L. Regulation of the vitamin D receptor gene by environment, genetics and epigenetics. Gene 561, 171-180,
doi:10.1016/j.gene.2015.02.024 (2015).
Pilon, C. et al. Methylation Status of Vitamin D Receptor Gene Promoter in Benign and Malignant Adrenal Tumors. Int J Endocrinol 2015, 375349, doi:10.1155/2015/375349 (2015).
Chandel, N., Malhotra, A. & Singhal, P. C. Vitamin D receptor and epigenetics in HIV infection and drug abuse. Front Microbiol 6, 788, doi:10.3389/fmicb.2015.00788 (2015).
Jiang, C. et al. The methylation state of VDR gene in pulmonary tuberculosis patients. J Thorac Dis 9, 4353-4357, doi:10.21037/jtd.2017.09.107 (2017).
Hussain, M. Z. et al. Genetic and expression deregulation of immunoregulatory genes in rheumatoid arthritis. Mol Biol Rep 48, 5171-5180, doi:10.1007/s11033-021-06518- 3 (2021).
Sun, J., Zhang, S., Liu, J. S., Gui, M. & Zhang, H. Expression of vitamin D receptor in renal tissue of lupus nephritis and its association with renal injury activity. Lupus 28, 290-294, doi:10.1177/0961203319826704 (2019).
Matos, C. et al. Downregulation of the vitamin D receptor expression during acute gastrointestinal graft versus host disease is associated with poor outcome after allogeneic stem cell transplantation. Frontiers in immunology 13, 1028850, doi:10.3389/fimmu.2022.1028850 (2022).
Neme, A., Seuter, S. & Carlberg , C. Vitamin D-dependent chromatin association of
CTCF in human monocytes. Biochim Biophys Acta 1859, 1380-1388, doi:10.1016/j.bbagrm.2016.08.008 (2016).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376-380, doi:10.1038/nature11082 (2012).
Genomes Project, C. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56-65, doi:10.1038/nature11632 (2012).
Rieder, M. J. et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med 352, 2285-2293, doi:10.1056/NEJMoa044503 (2005).
Carlberg , C. & Haq, A. The concept of the personal vitamin D response index. J Steroid Biochem Mol Biol 175, 12-17, doi:10.1016/j.jsbmb.2016.12.011 (2018).
Carlberg , C. et al. Primary vitamin D target genes allow a categorization of possible benefits of vitamin D3 ^supplementation. PLoS ONE 8, e71042, doi:10.1371/journal.pone.0071042 (2013).
Wilfinger, J. et al. Primary vitamin D receptor target genes as biomarkers for the vitamin D3 status in the hematopoietic system. J. Nutr. Biochem. 25, 875-884 (2014).
Ryynanen, J. et al. Changes in vitamin D target gene expression in adipose tissue monitor the vitamin D response of human individuals. Mol Nutr Food Res 58, 20362045, doi:10.1002/mnfr.201400291 (2014).
Saksa, N. et al. Dissecting high from low responders in a vitamin D3 intervention study. J Steroid Biochem Mol Biol 148, 275-282, doi:10.1016/jjsbmb.2014.11.012 (2015).
Vukic, M. et al. Relevance of vitamin D receptor target genes for monitoring the vitamin D responsiveness of primary human cells. PLoS ONE 10, e0124339, doi:10.1371/journal.pone.0124339 (2015).
Seuter, S. et al. Molecular evaluation of vitamin D responsiveness of healthy young adults. J Steroid Biochem Mol Biol 174, 314-321 (2017).
Mangin, M., Sinha, R. & Fincher, K. Inflammation and vitamin D: the infection connection. Inflamm Res 63, 803-819, doi:10.1007/s00011-014-0755-z (2014).
Salzer, J. et al. Vitamin D as a protective factor in multiple sclerosis. Neurology 79, 2140-2145, doi:10.1212/WNL.0b013e3182752ea8 (2012).
Fleet, J. C., DeSmet, M., Johnson, R. & Li, Y. Vitamin D and cancer: a review of molecular mechanisms. Biochem J 441, 61-76, doi:10.1042/BJ20110744 (2012).
Jiang, X. et al. Genome-wide association study in 79,366 European-ancestry individuals informs the genetic architecture of 25-hydroxyvitamin D levels. Nature communications 9, 260, doi:10.1038/s41467-017-02662-2 (2018).
Prabhu, A. V., Luu, W., Li, D., Sharpe, L. J. & Brown, A. J. DHCR7: a vital enzyme switch between cholesterol and vitamin D production. Prog Lipid Res 64, 138-151, doi:10.1016/j.plipres.2016.09.003 (2016).
Hanel, A., Veldhuizen, C. & Carlberg , C. Gene-regulatory potential of 25-hydroxyvitamin D3 and D2. Front Nutr 9, 910601, doi:10.3389/fnut.2022.910601 (2022).
Seuter, S., Pehkonen, P., Heikkinen, S. & Carlberg , C. Dynamics of 1a,25-dihydroxyvitamin D-dependent chromatin accessibility of early vitamin D receptor target genes. Biochim Biophys Acta 1829, 1266-1275, doi:10.1016/j.bbagrm.2013.10.003 (2013).
Kreienkamp, R. et al. Vitamin D receptor signaling improves Hutchinson-Gilford progeria syndrome cellular phenotypes. Oncotarget, 30018-30031, doi:10.18632/oncotarget. 9065 (2016).
Manson, J. E. et al. Vitamin D supplements and prevention of cancer and cardiovascular disease. N Engl J Med 380, 33-44, doi:10.1056/NEJMoa1809944 (2019).
Chen, Y., Michalak, M. & Agellon, L. B. Importance of nutrients and nutrient metabolism on human health. Yale J Biol Med 91, 95-103 (2018).
Ordovas, J. M., Ferguson, L. R., Tai, E. S. & Mathers, J. C. Personalised nutrition and health. Bmj 361, bmj k2173, doi:10.1136/bmj.k2173 (2018).
Carlberg , C. et al. In vivo response of the human epigenome to vitamin D: a proof-of– principle study. J Steroid Biochem Mol Biol 180, 142-148, doi:10.1016/j.jsbmb.2018.01.002 (2018).
Neme, A. et al. In vivo transcriptome changes of human white blood cells in response to vitamin D. J Steroid Biochem Mol Biol 188, 71-76, doi:10.1016/jjsbmb.2018.11.019 (2019).
Verstuyf, A., Carmeliet, G., Bouillon, R. & Mathieu, C. Vitamin D: a pleiotropic hormone. Kidney international 78, 140-145 (2010).
Carlberg , C. The physiology of vitamin D-far more than calcium and bone. Front Physiol 5, 335, doi:10.3389/fphys.2014.00335 (2014).
See also by Carlberg in Vitamin D Life
Vitamin D Nutrigenomics - High, Medium, and Low Responders - March 2019
Vitamin D in the Context of Evolution (400,000,000 years) – July 2022
6 of 6,980 results for nutrigenomics "vitamin d" in Google Scholar (Oct 2024)
Nongenomic Activities of Vitamin D - Dec 2022 FREE PDF
Using nutrigenomics to guide personalized nutrition supplementation for bolstering immune system - Jan 2023 c10.1007/s13755-022-00208-5|FREE PDF]
Nutrigenomics in Autoimmune Disease - 2024 Nutrigenomics in Autoimmune Disease - 2024 https://doi.org10.4018/979-8-3693-5528-2.ch01
Intervention Approaches in Studying the Response to Vitamin D3 Supplementation- Carlberg- July 2023 FREE PDF
Methylation of the Vitamin D Receptor Gene in Human Disorders - Dec 2023 FREE PDF
Vitamin D and Aging: Central Role of Immunocompetence- Carlberg - Jan 2024 FREE PDF
Vitamin D Life - Response to Vitamin D - many studies 149+
Vitamin D Life - Vitamin D Receptor activation can be increased in 14 ways
Omega-3, Magnesium, Zinc, Quercetin, non-daily Vit D, Curcumin, intense exercise, Butyrate Ginger, Essential oils, etc Note: The founder of Vitamin D Life uses 10 of the 14 known VDR activators
Vitamin D Life – The Vitamin D Receptor is associated with many health problems __
{include}
Vitamin D Life – Vitamin D Receptor category:
{include}
Vitamin D Life – Genetics category contains
{include}
